How Does the RB1 Protein Function?
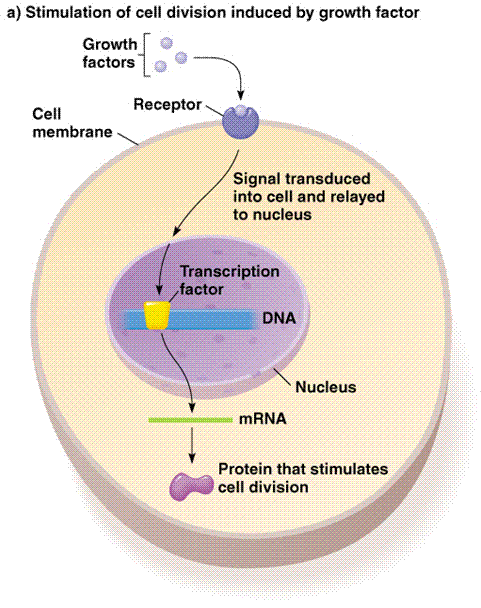
Let's first take a look at a typical, signal transduction pathway. This will give a general idea behind how proteins work to turn on and off the transcription of genes. This diagram shows exactly what we learned in class: environmental signal proteins stimulate a series of events that lead to a responce element entering the nucleus and connecting with a gene promoter. By doing this, transcription can either begin or cease, based on the particular signal.
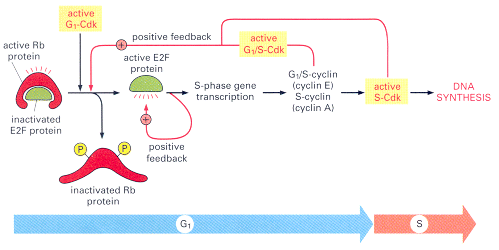
While the Retinoblastoma protein differs in its function, it acts in a similar manner to typical signal transduction proteins. Here's how it works. The RB1 gene is widely expressed, encoding a 110-kd (4.7 kb) nuclear protein, pRb. Generally speaking, in normal cells pRb is inactivated by phosphorylation and activated by dephosphorylation. Active (dephosphorylated) pRb binds and inactivates the cellular transcription factor E2F1, the function of which is required for cell cycle progression (14).
Going even further in detail, E2F binds to specific DNA sequences in the promoters of many genes that encode proteins required for S-phase entry, including G1/S-cyclins and S-cyclins. E2F's function is controlled primarily by an interaction with the retinoblastoma protein. During G1, Rb binds to E2F and blocks the transcription of S-phase genes. When cells are stimulated to divide by extracellular signals, active G1-Cdk accumulates and phosphorylates Rb, reducing its affinity for E2F. Phosphorylation is governed by a cascade of cyclins, cyclin-dependent kinases and cyclin kinase inhibitors The Rb then dissociates, allowing E2F to activate S-phase gene expression. This G1-S checkpoint seems to be the most crucial in the cell cycle, and thus the major source of cancer (14).
The complete loss of Rb does not immediately cause increased proliferation of other cell types, in part because Hct1 and p27 (two other proteins) provide assistance in G1 control, and in part because other cell types contain Rb-related proteins that provide backup support in the absence of Rb (14). It is also likely that other proteins, unrelated to Rb, help to regulate the activity of E2F.
The Size and Structure of Rb Protein
<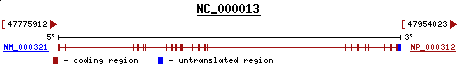 >
>
NM_000321 4740 bp mRNA linear
A 928 Amino Acid Protein.
It contains a putative "leucine-zipper" motif that is exclusively encoded by exon-20.
Rb A = Comprising 268 amino acids, this is the Retinoblastoma-associated protein A domain. This domain has the cyclin fold as predicted (14).
Rb B = Comprising 173 amino acids, this is the Retinoblastoma-associated protein B domain. The crystal structure of the Rb pocket bound to a nine-residue E7 peptide containing the LxCxE motif, shared by other Rb-binding viral and cellular proteins, shows that the LxCxE peptide binds a highly conserved groove on the B domain. The B domain has a cyclin fold (14).