Experimental
|
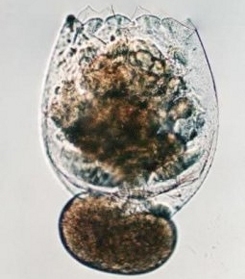
In the past few years, we have focused our research work on the homeobox genes and rRNA genes in invertebrates, particularly rotifers and cnidarians. We have been investigating these genes in these experimental organisms to study the molecular mechanisms of development, and the molecular evolution of complex gene families. Project (1): Molecular Evolution of the Hox, Pax, and Prospero gene families, and Molecular Mechanisms of Development as mediated by DNA:protein interactions in transcriptional networks
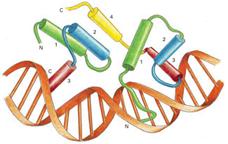
Why Homeobox Genes? These genes are critical for development in all eukaryotes, especially along the anterior-posterior axis during embryogenesis. The number and especially sequence variability of the Hox genes in a genome may be proportional to an organism's complexity. Hox genes act as "network I/O switches" to control the regulatory circuitry responsible for the patterning of spatial domains in developing embryos (from a "proximate" or mechanistic perspective), and ultimately for the morphological variation observed in phylogenetic histories (now from an "ultimate" or evolutionary perspective). Why Rotifers (and other creatures)? A large variety of shapes are found among the 2000 or so species of rotifers. A large variety of shapes are found among all ogranisms. Rotifers are microscopic invertebrates, and hence closer in evolutionary history to the ancestral progenitor of all metazoans. What determines Shape in organisms? What are the molecular mechanisms of not only development, but also of the processes that determine shape and form? How are these mechansims related? A very broad question: If evolution leads to adaptive complexity, as observed in the varied body-plans, physiologies, and biochemistries of all organisms, and if evolution also leads to diversification, as is evident from the millions of species of organisms, then what molecular mechanisms underlie these powerful forces of adaptation and diversification? In other words, what is the relationship between evolution and development? A narrower question: How are the cis-regulatory elements of BOTH transcription factor genes AND their target genes, which are "located" in the molecular subcircuits of interlocked networks, related to phenotypic variation? This last, more narrowly focused question can be addressed both computationally and experimentally for discrete "control-switch/subcircuit genes" combinations. Introduction Every eukaryotic organism has homeobox genes, which are typically identified by a conserved 3’ region of about 180 bp, encoding a carboxy-terminal 60 amino acid DNA-binding domain consisting of 3 or sometimes 4 alpha helices in the classical HTH (helix-turn-helix) motif. The homeobox genes provide morphological identity to clusters of embryonically derived cells programmed with positional information by the homeobox gene products acting as TFs (transcription factors), most often for anterior-posterior development during embryogenesis. The exact developmental mechanisms and pathways of these complex processes are not understood fully for development, and especially for shape. Two broad questions emerge: How does a single zygotic cell develop into a fully functional organism with multiple tissues and organs? What determines the final shapes of organisms (a rotifer or an insect or a mammal) and the parts of bodies (a hand or a foot, a mouth or an eye)? Because of the highly conserved amino acid sequence in the DNA binding helix (3 rd helix) homeobox genes also provide insights into phylogenetic relationships. We can study these relationships by the use of both sequence and structural homology methods. The techniques of Bioinformatics are especially important in this regard. One goal in this regard is to view the Tree of Life through the "lens" of DNA:protein bindings within regulatory networks, and to discover an underlying pattern of relationships that helps to define and clarify gene phyolgenies. Some older classification schemes placed the rotifers next to the nematodes as "cousins", and therefore implied that the two organisms had highly homologous homeobox genes. However, the rotifers in general develop a significantly more morphologically complex body than the nematodes. This phenotypic expression of the genes in question suggests that the genes may not be so closely homologous, or if it were the case that the genes in question were highly homologous and the two organism’s morphologies noticeably different, then the genes' expression must be regulated differently in each organism. Recent phylogenetic analyses from both rRNA genes and Hox genes place the nematodes near Arthropods (Drosophila) on the Tree of Life, whereas the rotifers are now near Platyhelminthes, in the newly defined Lophotrochozoans. The Tree of Life as we humans know it is contantly "evolving" with the more recent molecular analyses and results. The Fornari lab is interested in (and see Research Statement): 1. Structure of the helix-turn-helix motif (HTH), Homeodomain (HD), Prospero (Prox), and paired (Pax) domains in Transcription Factors and other regulatory proteins The use of powerful modern molecular modeling programs, such as Chimera and Deepview, in conjunction with the online protein databases allows for efficient analysis of the physical properties of proteins and their biochemistry at the molecular level, including DNA binding and protein-protein interactions. 2. Molecular Mechanisms and Function of Homeobox Genes/Proteins Traditional methods from biochemistry and molecular biology, such as PCR, cloning, and DNA sequencing, and gene expression, are used to study the structure/function relationship of the homeobox genes/proteins, and their regulatory mechanisms. 3. Molecular Evolution of the HTH, Prospero, Homeobox, and Pax Genes The use of tools in the realm of bioinformatics allows for (1) the investigation of evolutionary relationships, (2) the discovery of new knowledge about homeodomain structure and function, and (3) the incorporation of new knowledge into phylogenetic models of evolution and development. 4. Method Development Experimental molecular biology, biochemistry, and bioinformatics are applied to rotifers, cnidarians, sponges, and other organisms to elucidate and evaluate nucleic acid and protein sequences. State of the art automated DNA sequencing by a Beckman-Coulter CEQ 8000 Genetic Analysis System is employed to determine and evaluate sequence data. Version 2.0 |