

What is gene linkage?
What initial linkage studies to isolate the hMLH1
gene were done?
What further linkage analysis has been done?
Chromosomes can be divided up into discrete sections of DNA using restriction fragment length polymorphisms. This means that same discrete sections of DNA in one individual can be cut into two fragments while the section in another individual can not be cut into two fragments. See figure for further explanation.
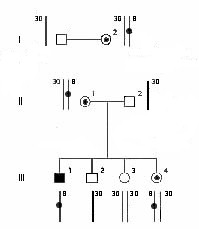 |
In the figure to the left, the known marker is represented by the chromosome with the circle. This marker can be inherited from one of the parents. The inheritance of this marker with the disease phenotype are compared and if there is a high probablility of co-inheritance (such as 99%), then the disease gene (its exact location is not known) and the marker are said to be linked. The marker is in a known position, so the disease gene can be pinpointed to somewhere close to this marker. |
An individual inherits one of each chromosome from each parent. During meiosis, pairs of homologous chromosomes come together and may exchange segments in a process known as recombination. Recombination results in the switching of segments between homologous chromosomes.
Recombination plays an important part of linkage analysis because the percentage of instances that a know marker (i.e. a section that is cut in one fragment, but not in one from another individual) is inherited with a disease gene can be used to find how close the marker and gene are found in the genome. The closer a marker is to a disease gene the higher the percentage of co-inheritance because there is a smaller amount of space for recombination to occur between the genes.
What initial linkage studies to isolate the hMLH1 gene were done?
Initially Hereditary non-polyposis colon cancer (HNPCC) was linked to chromosome 2; however, not all cases of clinically diagnosed HNPCC could be liked to this chromosome. Three Families that fit this case were first analyzed for linkage of two known hereditary colon cancer genes (APC and DCC genes). The unknown disease gene segregated separately from the genes on chromosome 2, therefore demonstrating that these genes were not the cause of these cases of colon cancer (Ref. 5).
 |
Using family trees (pedigrees) that list individuals
affected with colon cancer and comparing DNA among as many individuals
listed in each of the family trees, co-inheritance of a known marker with
the disease phenotype was found. A total of 45 markers (RFLP’s) were
tested before segregation consistent with linkage was found on chomosome
3p21. Two of the three families showed linkage to this region and
the third did not. Of the two families that did show linkage, markers
closely located to the HNPCC gene on chromosome 2 excluded linkage in one
family and had a very low linkage on the second family.
Using other markers closely located to the original known marker, a locus on chromosome 3p21 - 23 was found to predispose to HNPCC. |
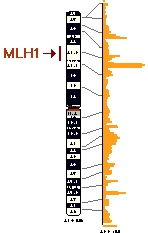
In further studies of the HNPCC gene on the chromosome
3, somatic cell hybridization was used to localize hMLH1 to this chromosome.
The hMLH1 gene is a yeast mismatch repair gene and experiments suggested
that HNPCC was associated with a defect in an early step of mismatch repair.
Through fluorescence in situ hybridization, the gene was isolated to the
3p21.3 region and was isolated further between two known markers (D3S1298
and D3S1561) by physically mapping the region and performing linkage anaylsis
(Ref. 14).
|